Our latest publications:
Lorena González, Lucía Díaz, Joan Pous, Blazej Baginski, Anna Duran-Corbera, Margherita Scarpa, Isabelle Brun-Heath, Ana Igea, Pau Martin-Malpartida, Lidia Ruiz, Chiara Pallara, Mauricio Esguerra, Francesco Colizzi, Cristina Mayor-Ruiz, Ricardo M Biondi, Robert Soliva, Maria J Macias, Modesto Orozco, Angel R Nebreda Nature Communications volume 14, Article number: 3318 (2023), doi:10.1038/s41467-023-39051-x.
p38α is a versatile protein kinase that can control numerous processes and plays important roles in the cellular responses to stress. Dysregulation of p38α signaling has been linked to several diseases including inflammation, immune disorders and cancer, suggesting that targeting p38α could be therapeutically beneficial. Over the last two decades, numerous p38α inhibitors have been developed, which showed promising effects in pre-clinical studies but results from clinical trials have been disappointing, fueling the interest in the generation of alternative mechanisms of p38α modulation. Here, we report the in silico identification of compounds that we refer to as non-canonical p38α inhibitors (NC-p38i). By combining biochemical and structural analyses, we show that NC-p38i efficiently inhibit p38α autophosphorylation but weakly affect the activity of the canonical pathway. Our results demonstrate how the structural plasticity of p38α can be leveraged to develop therapeutic opportunities targeting a subset of the functions regulated by this pathway.
Radoslaw Pluta, Eric Aragón, Nicholas A. Prescott, Lidia Ruiz, Rebeca A. Mees, Blazej Baginski, Julia R. Flood, Pau Martin-Malpartida, Joan Massagué, Yael David, Maria J. Macias Nature Communications volume 13, Article number: 7279 (2022), doi:10.1038/s41467-022-34925-y.
Forkhead box H1 (FoxH1) is an essential maternal pioneer factor during embryonic development that binds to specific GG/GT-containing DNA target sequences. Here we have determined high-resolution structures of three FoxH1 proteins (from human, frog and fish species) and four DNAs to clarify the way in which FoxH1 binds to these sites. We found that the protein-DNA interactions extend to both the minor and major DNA grooves and are thus almost twice as extensive as those of other FOX family members. Moreover, we identified two specific amino acid changes in FoxH1 that allowed the recognition of GG/GT motifs. Consistent with the pioneer factor activity of FoxH1, we found that its affinity for nucleosomal DNA is even higher than for linear DNA fragments. The structures reported herein illustrate how FoxH1 binding to distinct DNA sites provides specificity and avoids cross-regulation by other FOX proteins that also operate during the maternal-zygotic transition and select canonical forkhead sites.
Pau Martin-Malpartida, Silvia Arrastia-Casado, Josep Farrera-Sinfreu, Rudolf Lucas, Hendrik Fischer, Bernhard Fischer, Douglas C.Eaton, Susan Tzotzos, Maria J.Macias
Computational and Structural Biotechnology Journal,Volume 20, 2022, Pages 2082-2090, doi:10.1016/j.csbj.2022.04.031
Tumor necrosis factor (TNF) is a homotrimer that has two spatially distinct binding regions, three lectin-like domains (LLD) at the TIP of the protein and three basolaterally located receptor-binding sites, the latter of which are responsible for the inflammatory and cell death-inducing properties of the cytokine. Solnatide (a.k.a. TIP peptide, AP301) is a 17-mer cyclic peptide that mimics the LLD of human TNF which activates the amiloride-sensitive epithelial sodium channel (ENaC) and, as such, recapitulates the capacity of TNF to enhance alveolar fluid clearance, as demonstrated in numerous preclinical studies. TNF and solnatide interact with glycoproteins and these interactions are necessary for their trypanolytic and ENaC-activating activities. In view of the crucial role of ENaC in lung liquid clearance, solnatide is currently being evaluated as a novel therapeutic agent to treat pulmonary edema in patients with moderate-to-severe acute respiratory distress syndrome (ARDS), as well as severe COVID-19 patients with ARDS. To facilitate the description of the functional properties of solnatide in detail, as well as to further target-docking studies, we have analyzed its folding properties by NMR. In solution, solnatide populates a set of conformations characterized by a small hydrophobic core and two electrostatically charged poles. Using the structural information determined here and also that available for the ENaC protein, we propose a model to describe solnatide interaction with the C-terminal domain of the ENaCα subunit. This model may serve to guide future experiments to validate specific interactions with ENaCα and the design of new solnatide analogs with unexplored functionalities.
Our primary research interests are centered on deciphering intricate mechanisms that bridge cell signaling and gene expression, comprehending the regulation of these essential processes, and exploring their profound implications in a spectrum of human diseases.
In our laboratory, we seamlessly integrate cutting-edge molecular biology techniques with state-of-the-art tools, including NMR, X-ray, and SAXS, alongside the power of bioinformatics. These methods illuminate the structural intricacies of these molecular complexes. Furthermore, we have incorporated cryoEM into our array of structural biology techniques to delve into the study of expansive and multifaceted systems.
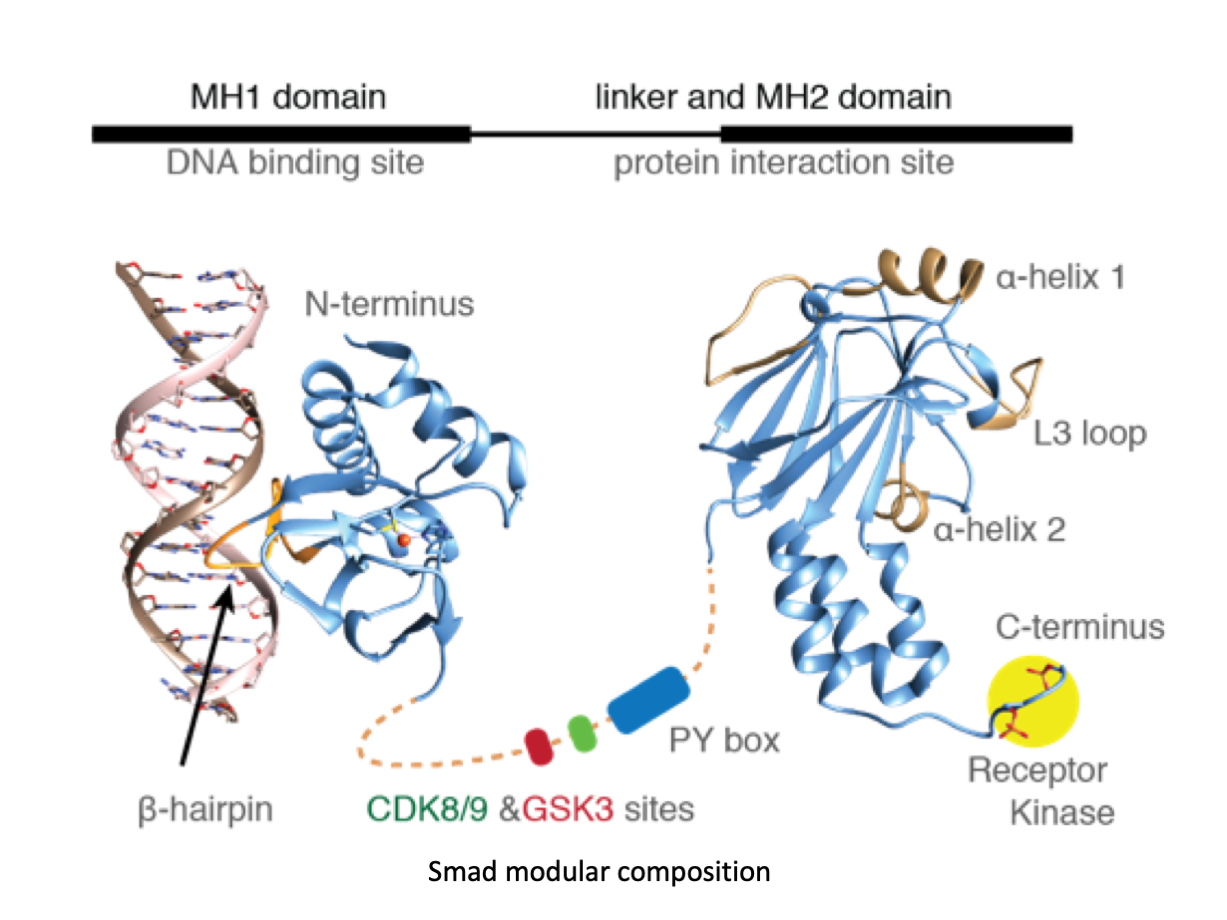
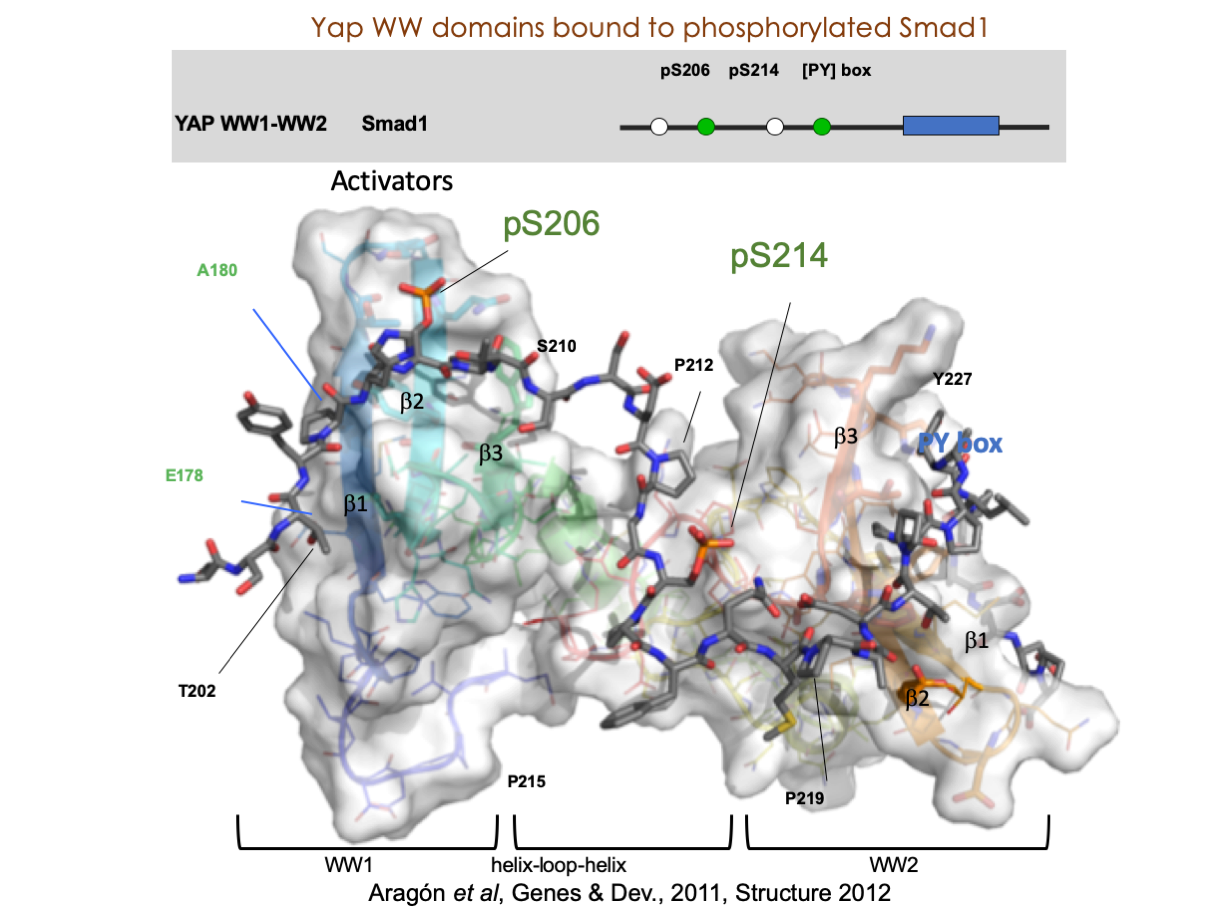
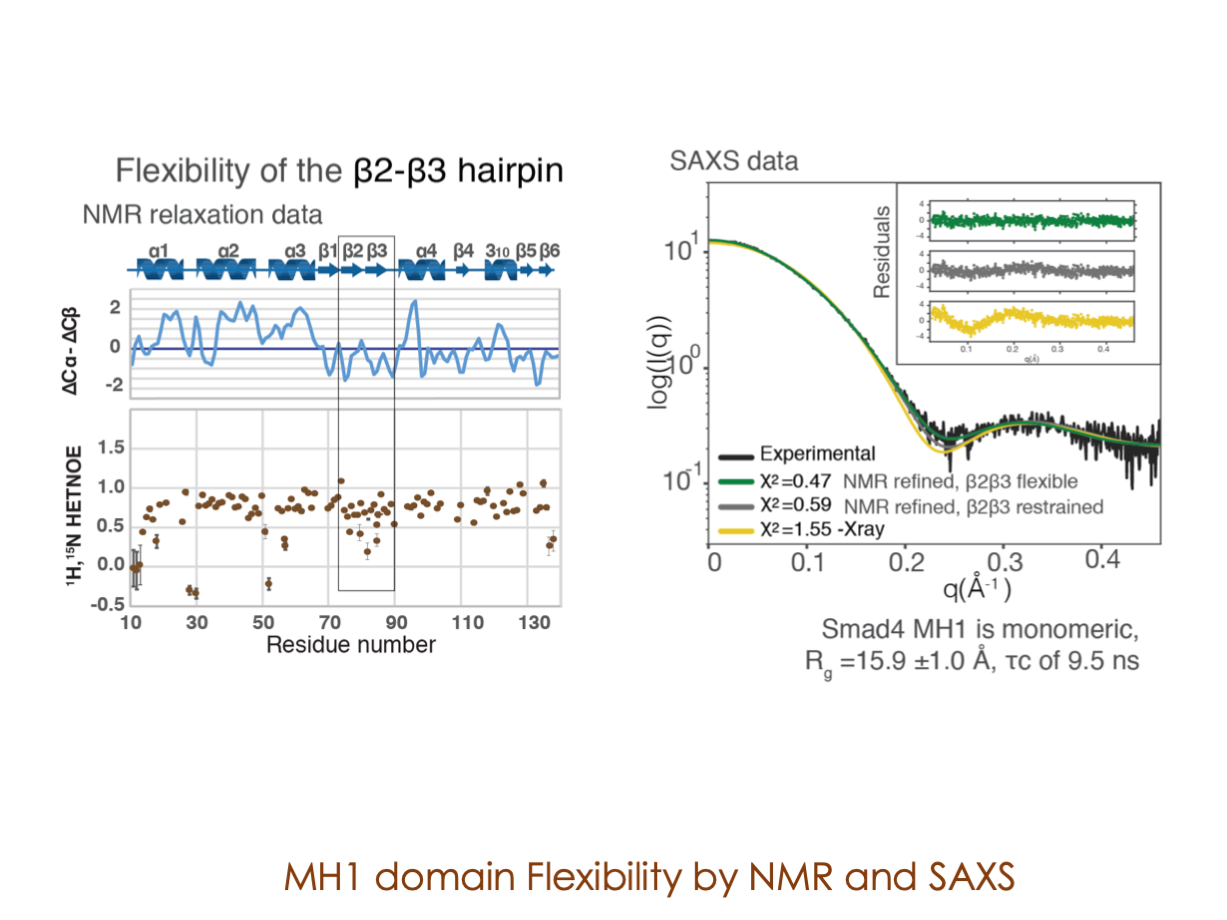
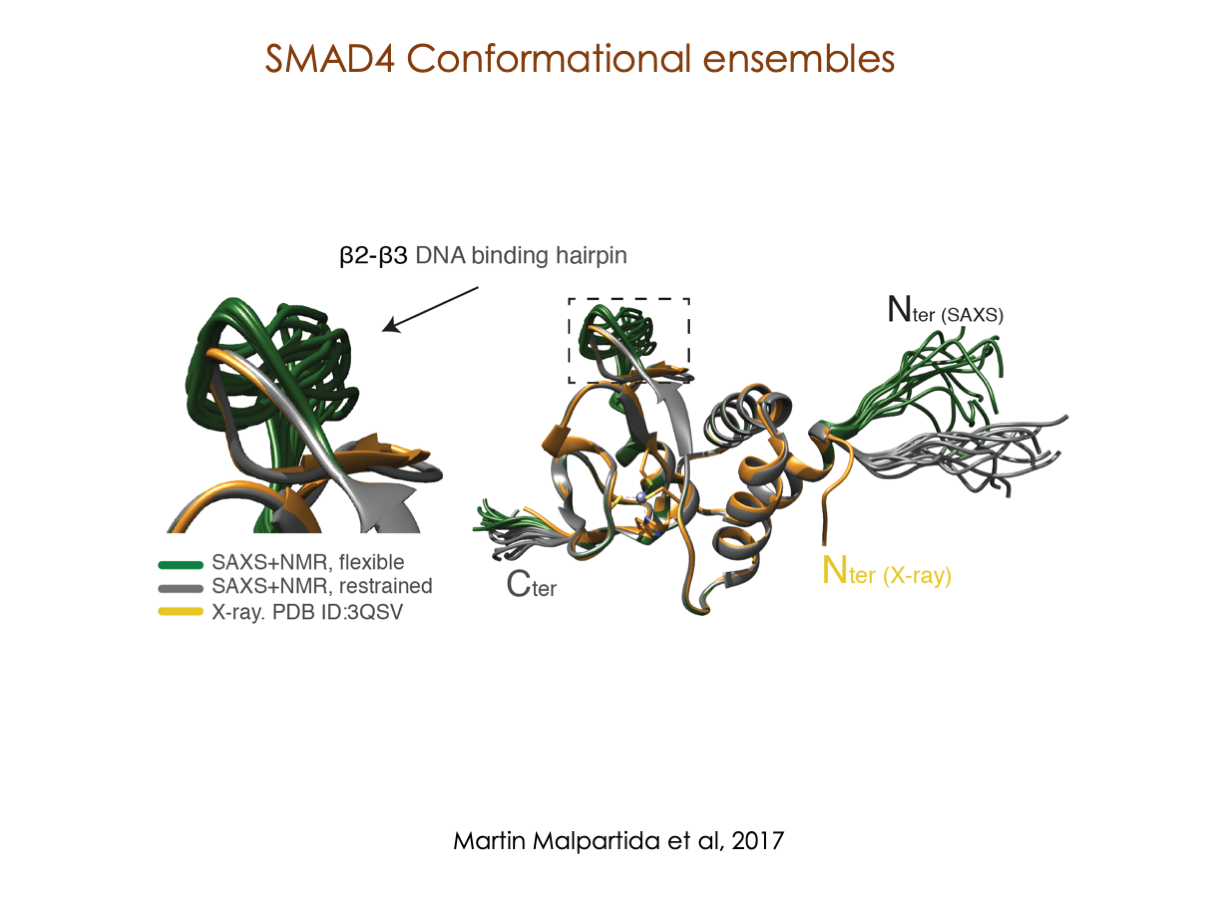
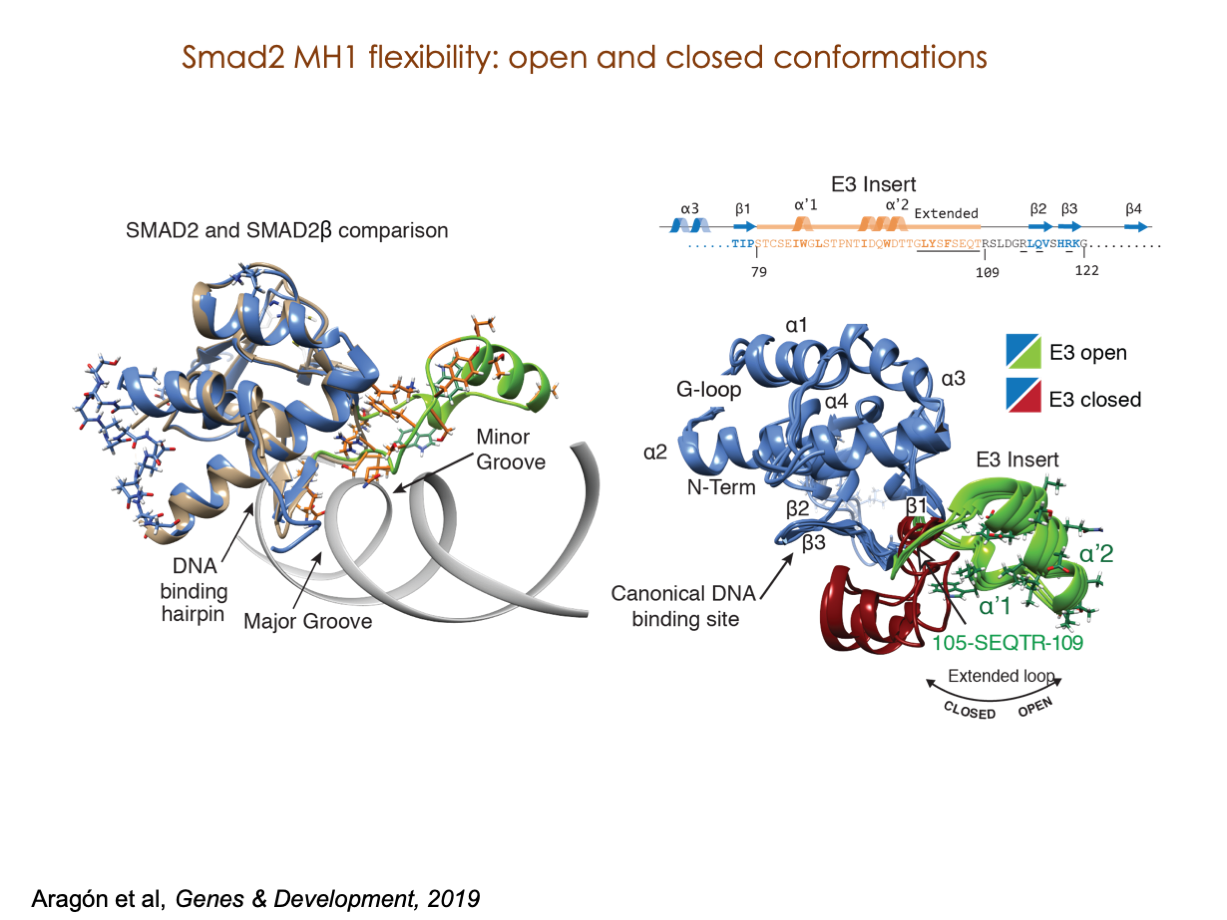
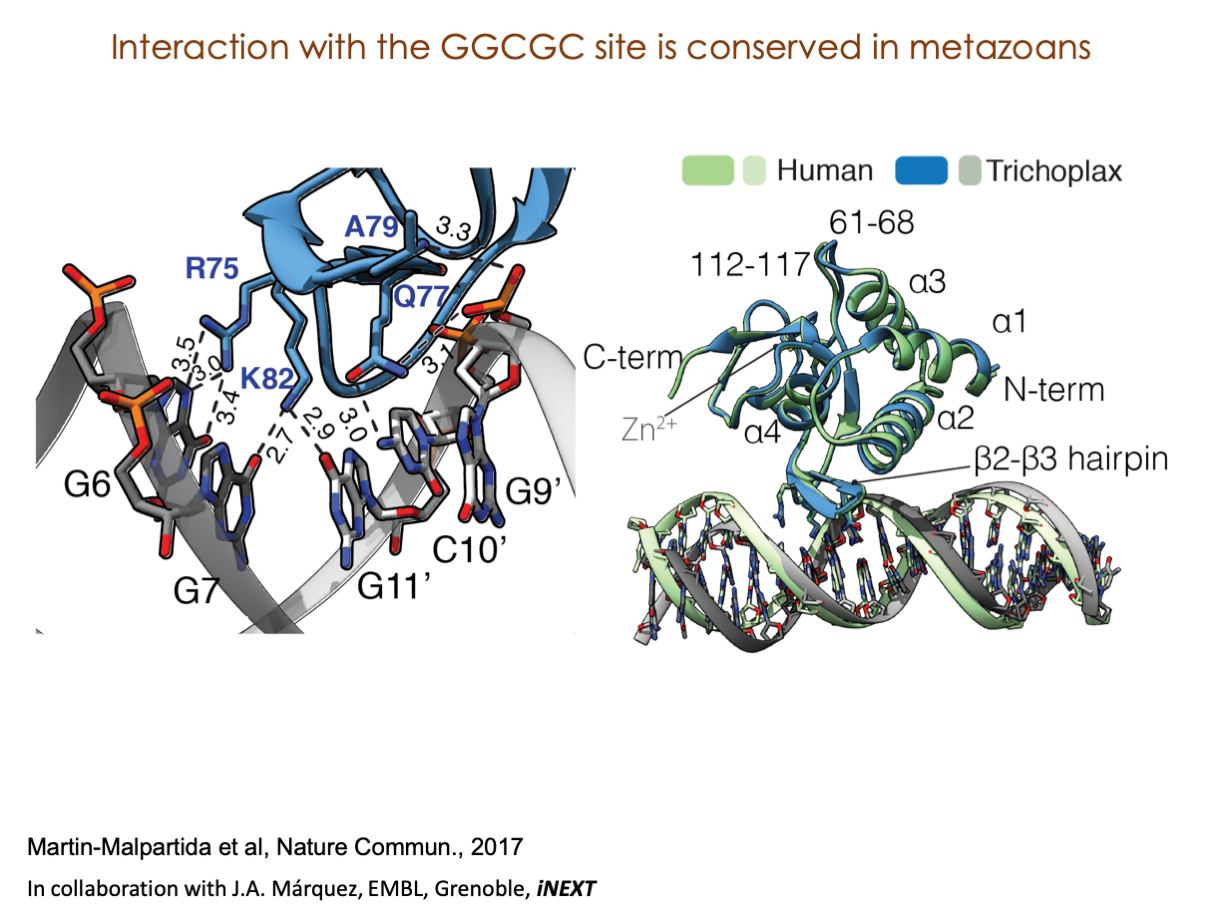
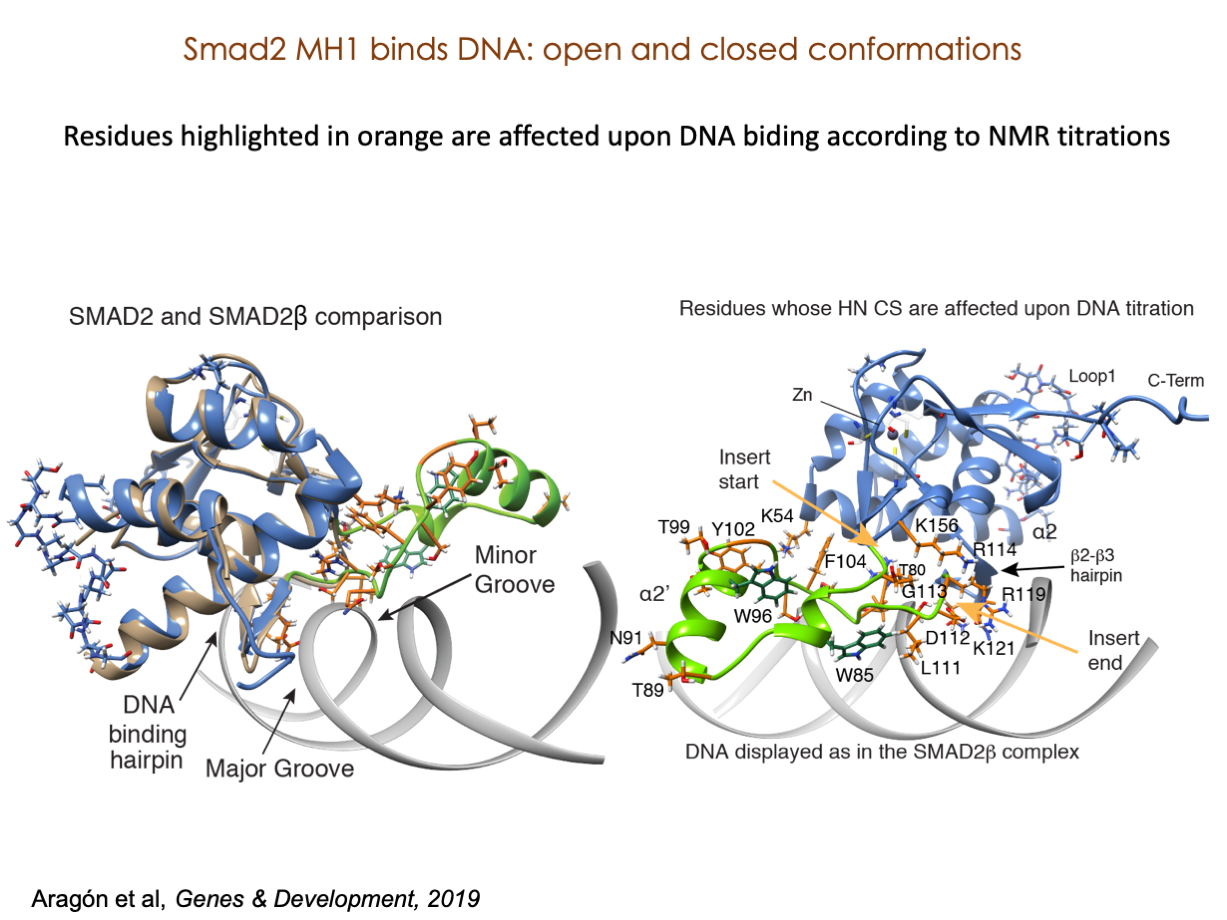
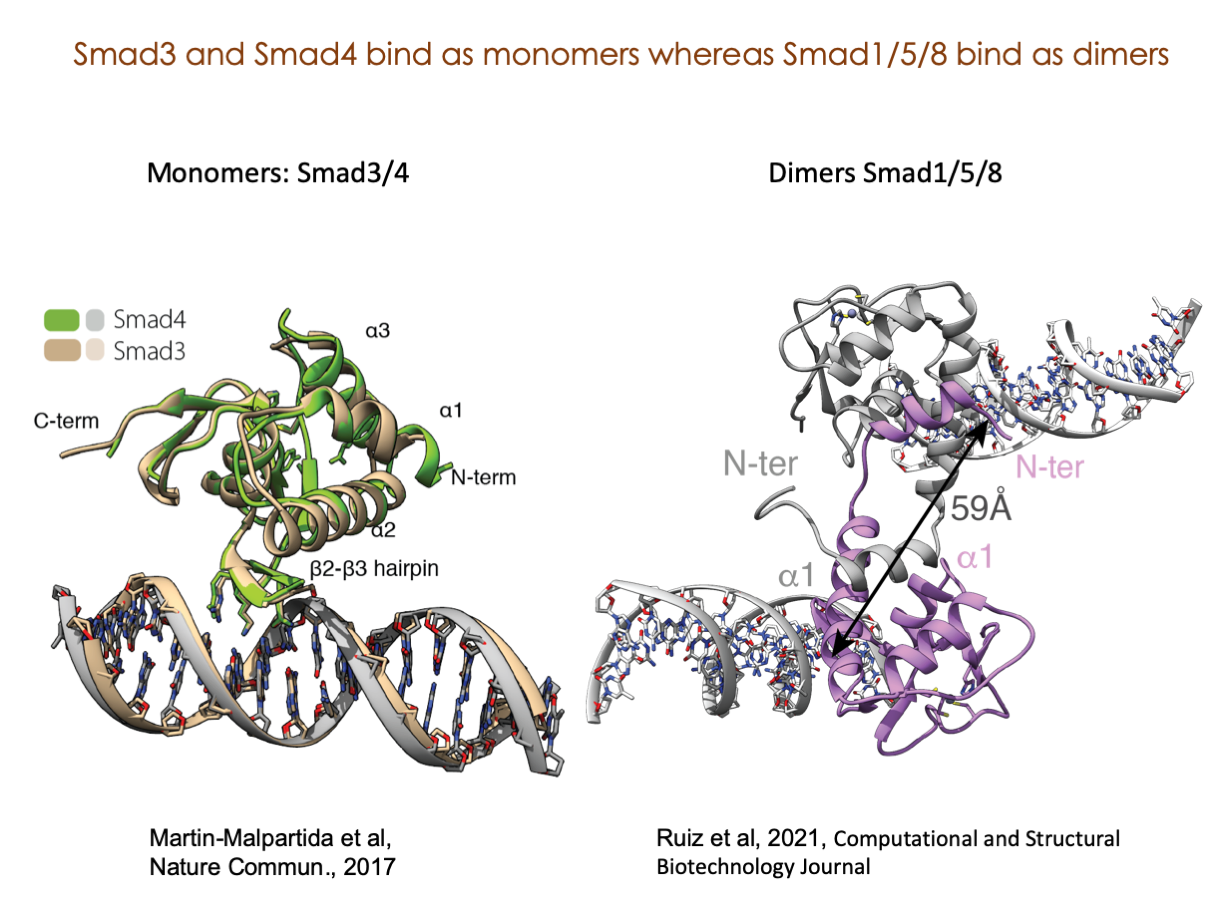
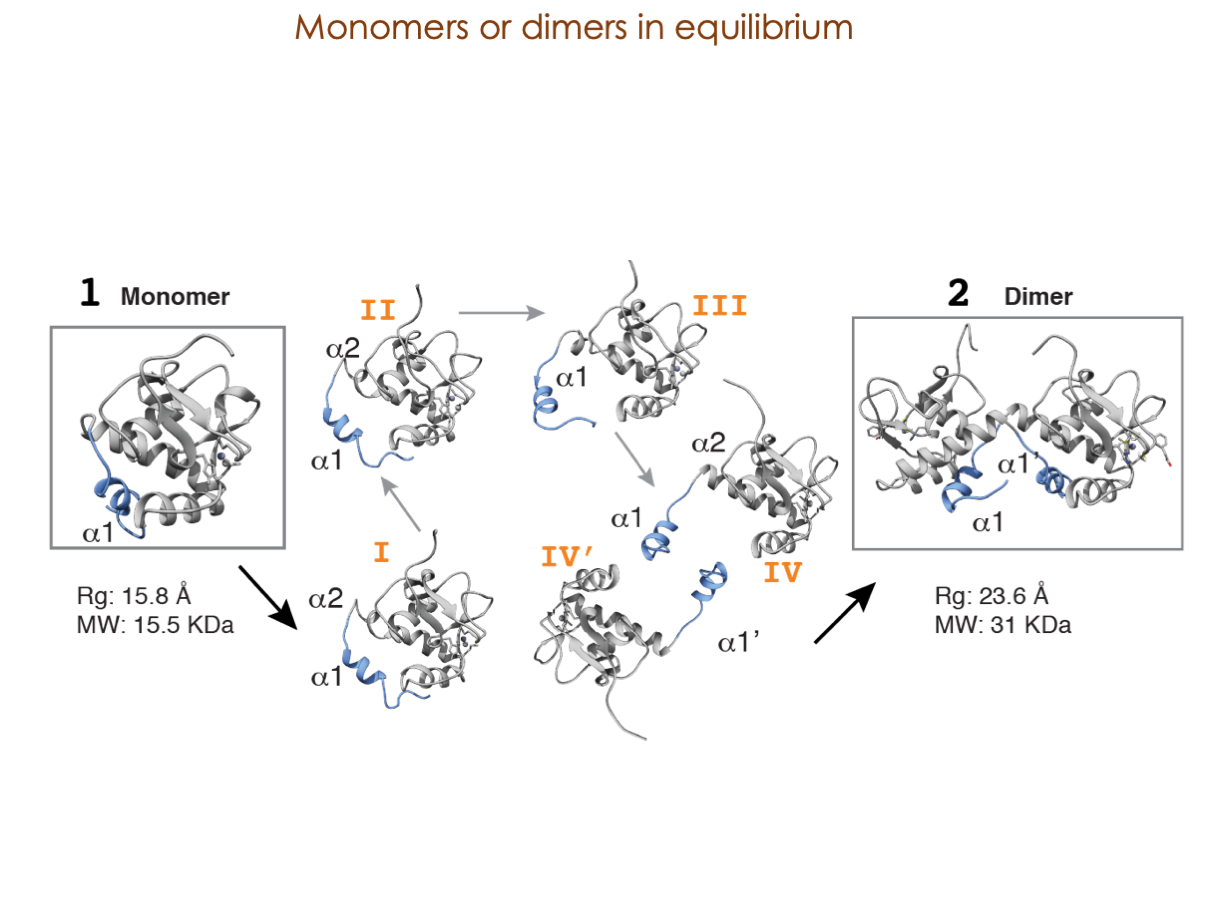
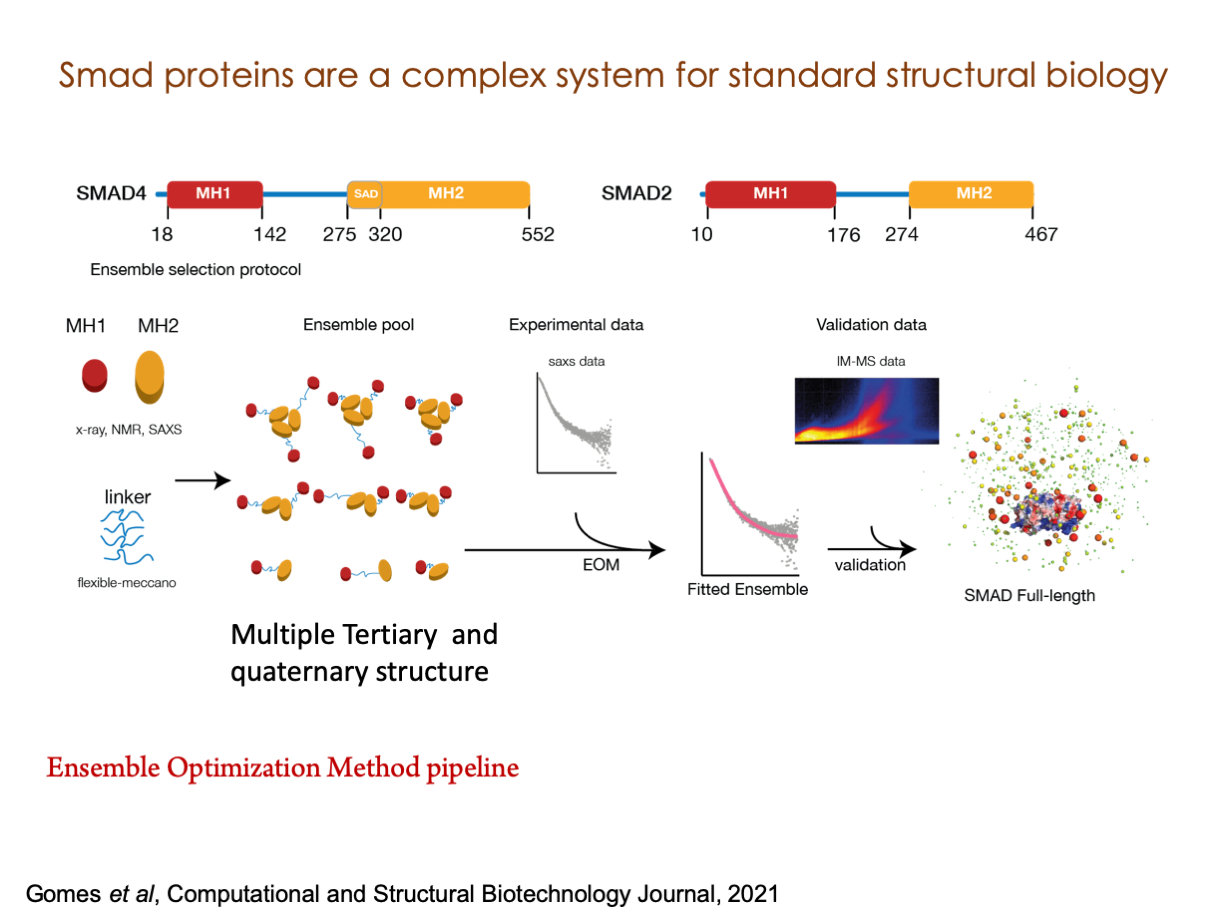
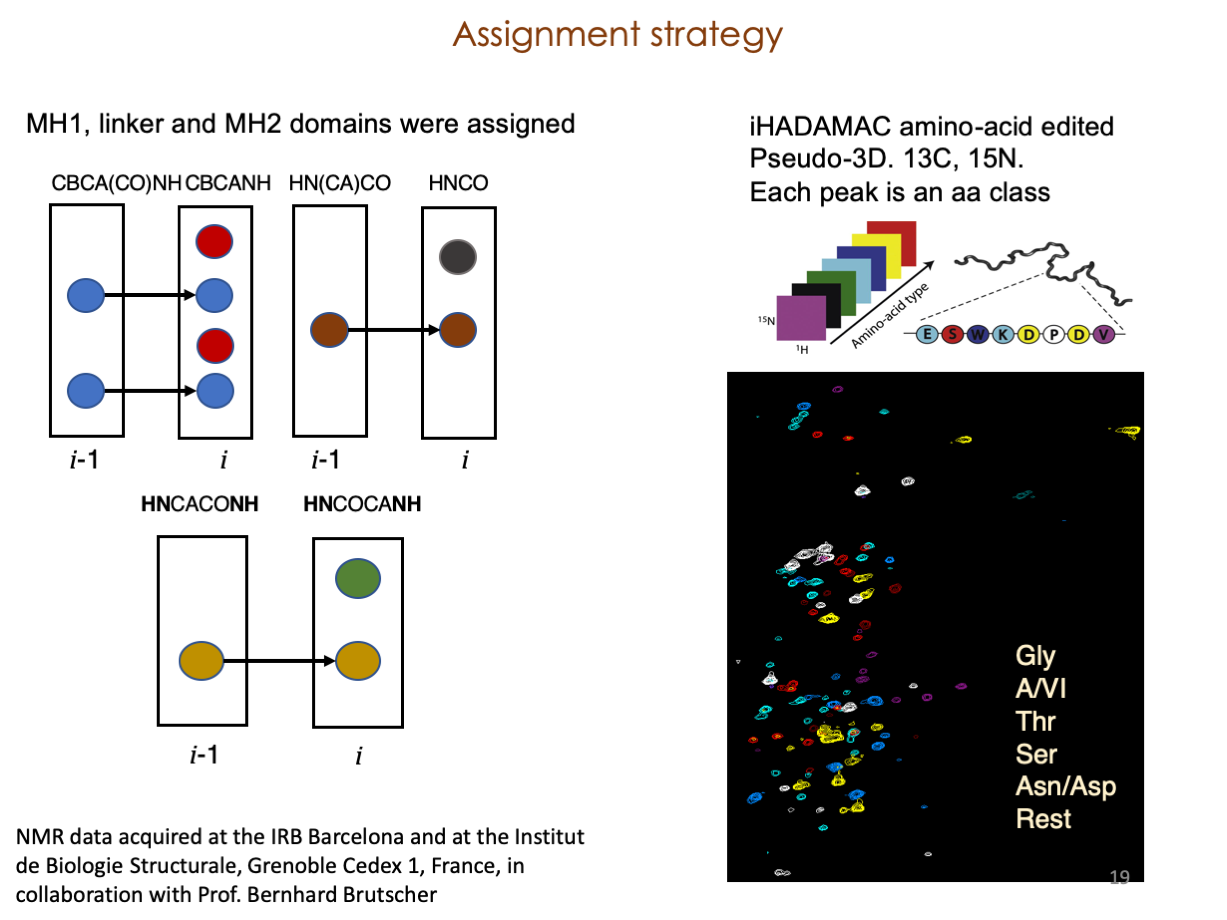
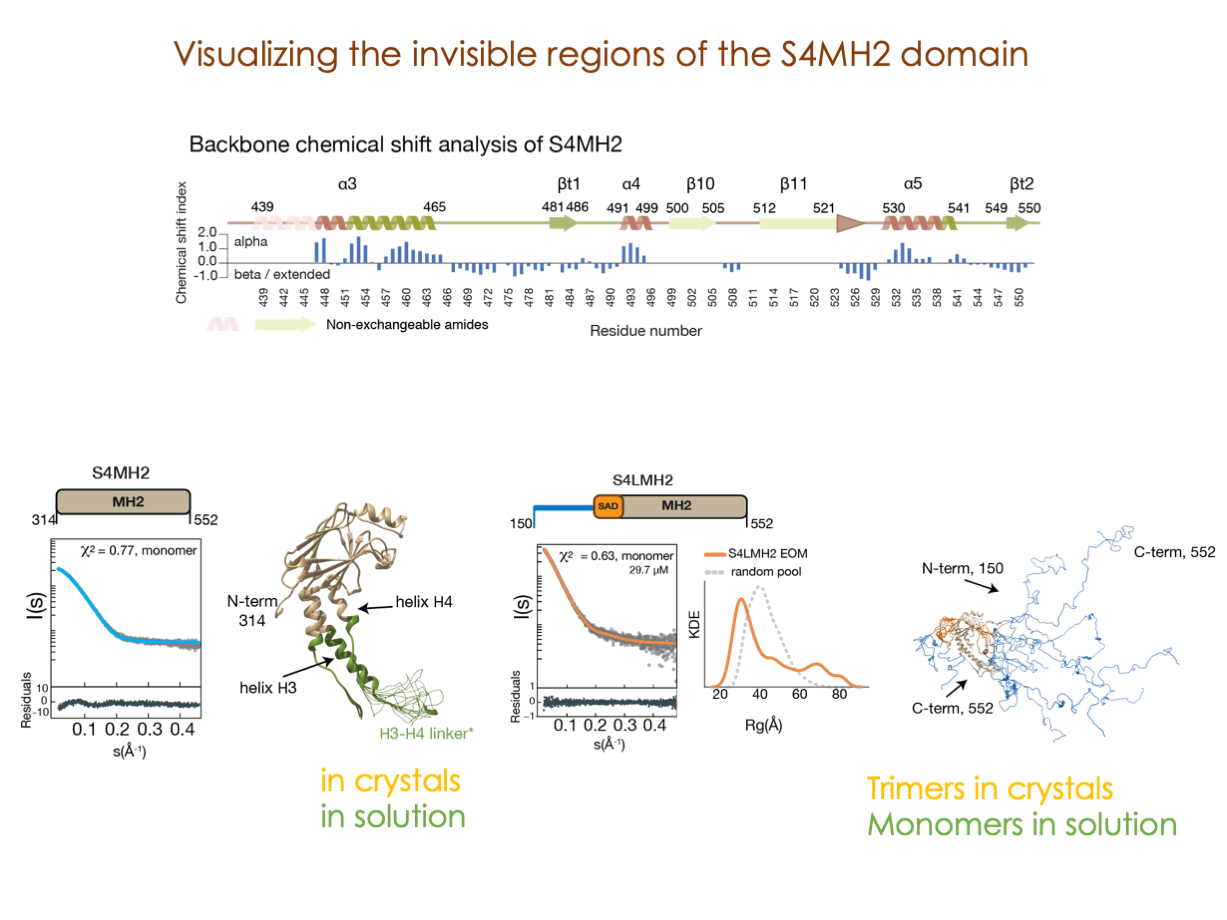
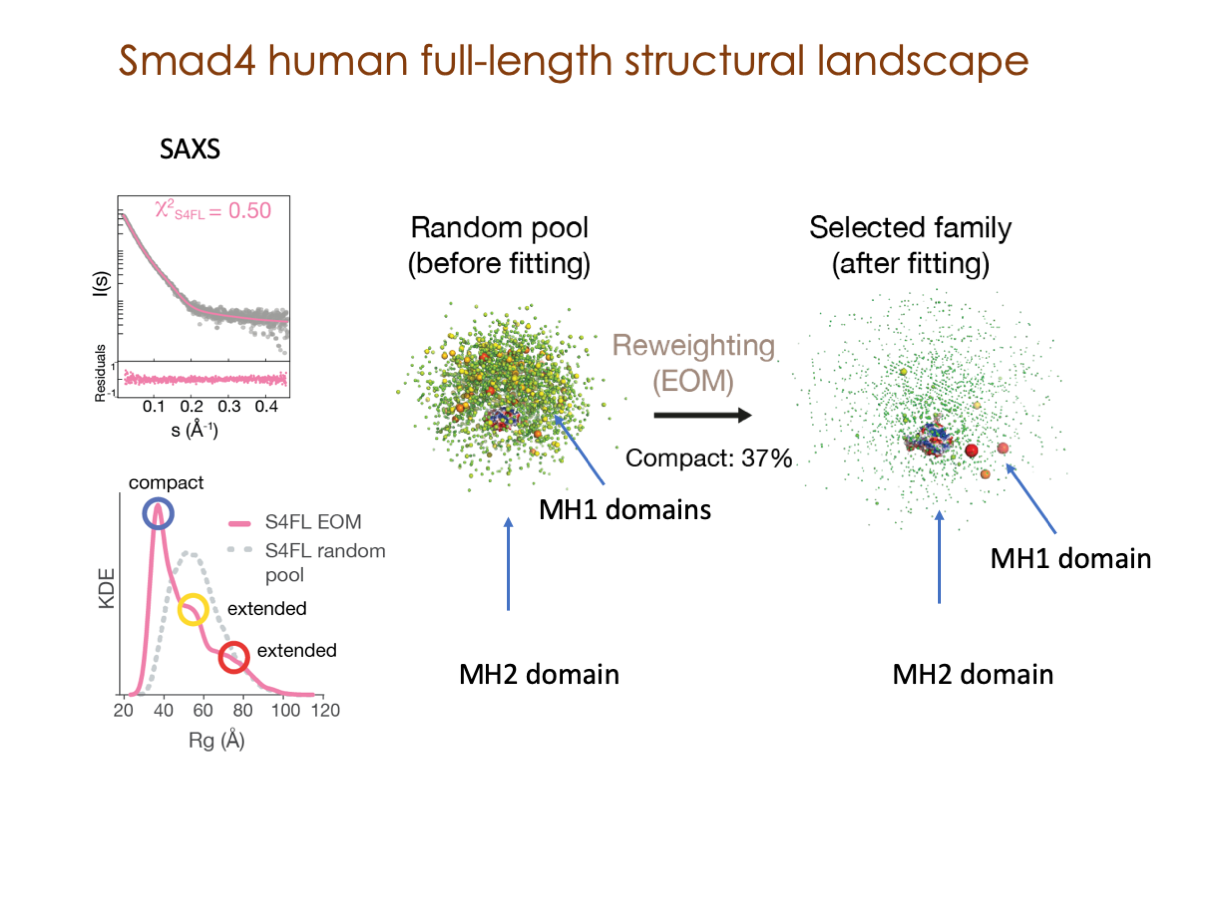
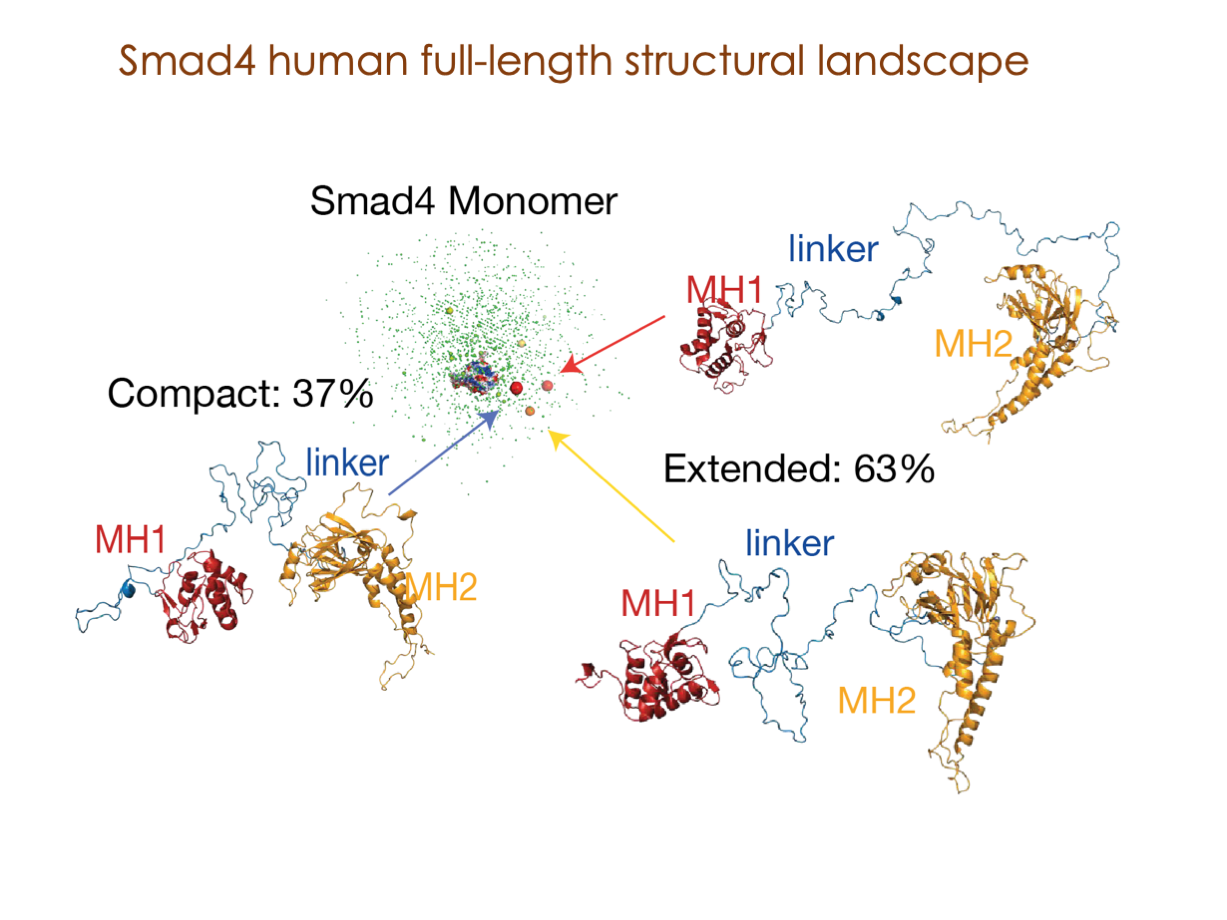
A significant portion of our research is dedicated to elucidating the nuances of TGFβ signaling, with a particular focus on the pivotal roles played by SMAD transcription factors and their associated cofactors. TGFβ signaling is integral to early embryogenesis and developmental processes. Remarkably, numerous malignancies employ analogous strategies to acquire aggressive attributes, thus enhancing their invasive potential and metastatic propensities.
Since 2008, our fruitful collaboration with the esteemed Joan Massagué’s group at the Memorial Sloan Kettering Cancer Center in New York, USA, has been concentrated on various facets of TGFβ signaling and SMAD functionality.
Moreover, in close collaboration with dedicated gynecologic and oncology medical professionals at Sant Pau Hospital in Barcelona, led by Ramon Rovira and colleagues, along with researchers at its affiliated research institute, we are diligently working to identify innovative diagnostic tools and predictive models for assessing the likelihood of relapse and metastasis in cases of Endometrial cancers.